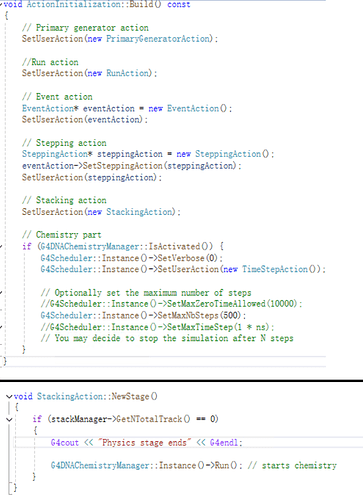
If I set like G4Scheduler::Instance()->SetMaxNbSteps(500); the program will end with segmentation fault (core dumped) after runing several protons. However, this problem will be solved when I set G4Scheduler::Instance()->SetMaxNbSteps(100) (at least it work out with 500 protons as beam). I know that this process controls the amount of chemcial reaction, however, 100 is clearly not enough in my case. I wonder what situation can possibly cause this? These are parts of snapshot from ActionInitialization.cc and Stacking Action.cc. Thanks if anyone could answer this.
#include “PhysicsList.hh”
#include “G4SystemOfUnits.hh”
#include “G4EmDNAChemistry.hh”
#include “G4EmDNAChemistryForPlasmids.hh” // Ensure this is included
#include “G4EmDNAPhysics.hh” // Include for the main physics option
#include “G4EmDNAPhysics_option2.hh” // Include for the second physics option
PhysicsList::PhysicsList(const G4String& option)
: G4VModularPhysicsList(),
fEmDNAPhysicsList(nullptr),
fEmDNAChemistryList(nullptr),
fEmDNAChemistryForPlasmids(nullptr), // Initialize the custom pointer
fPhysDNAName(“”)
{
G4double currentDefaultCut = 1. * nanometer;
G4ProductionCutsTable::GetProductionCutsTable()->SetEnergyRange(100 * eV, 1 * GeV);
SetDefaultCutValue(currentDefaultCut);
SetVerboseLevel(1);
// Choose physics list based on option
if (option == "G4EmDNAPhysics") {
fEmDNAPhysicsList = new G4EmDNAPhysics(); // Using option 2
}
else {
fEmDNAPhysicsList = new G4EmDNAPhysics_option2(); // Default option
}
RegisterConstructor("G4EmDNAChemistry"); // Register the new chemistry
}
PhysicsList::~PhysicsList()
{
delete fEmDNAPhysicsList; // Clean up the selected physics list
delete fEmDNAChemistryList;
delete fEmDNAChemistryForPlasmids; // Clean up the custom chemistry class
}
void PhysicsList::ConstructParticle()
{
if (fEmDNAPhysicsList) {
fEmDNAPhysicsList->ConstructParticle();
}
if (fEmDNAChemistryList) {
fEmDNAChemistryList->ConstructParticle();
}
if (fEmDNAChemistryForPlasmids) { // Use the custom chemistry class
fEmDNAChemistryForPlasmids->ConstructParticle();
}
}
void PhysicsList::ConstructProcess()
{
AddTransportation();
if (fEmDNAPhysicsList) {
fEmDNAPhysicsList->ConstructProcess();
}
if (fEmDNAChemistryList) {
fEmDNAChemistryList->ConstructProcess();
}
if (fEmDNAChemistryForPlasmids) { // Use the custom chemistry class
fEmDNAChemistryForPlasmids->ConstructProcess();
}
}
void PhysicsList::RegisterConstructor(const G4String& name)
{
if (name == fPhysDNAName) { return; }
if (verboseLevel > 0) {
G4cout << "===== Register constructor ==== " << name << G4endl;
}
if (name == "G4EmDNAChemistry") {
if (fEmDNAChemistryList) { return; }
fEmDNAChemistryList = new G4EmDNAChemistry();
fEmDNAChemistryList->SetVerboseLevel(verboseLevel);
}
else if (name == "G4EmDNAChemistryForPlasmids") { // Registering the custom chemical module
if (fEmDNAChemistryForPlasmids) { return; }
fEmDNAChemistryForPlasmids = new G4EmDNAChemistryForPlasmids(); // New instance
fEmDNAChemistryForPlasmids->SetVerboseLevel(verboseLevel);
G4cout << "Registered custom chemistry for plasmids" << G4endl;
}
else {
G4cout << "PhysicsList::RegisterConstructor: <" << name << ">"
<< " fails - name is not defined" << G4endl;
}
}
Is it because I have problems in writing physicslist.cc?
I tried the same thing with chem2 example, with 500 0.1 MeV protons, same problem occurs-segmentation fault (core dumped), it seems it cannot deal with many rounds of chemcial reactions?
Is it because step-by-step method is not allowed to do this amount of calculation?
Thanks, it seems about step-by-step method, in chem2 example, if you add more proton beams as resource, same problem will occur, but IRT method work out (with much lower calculation price), so I switch to this.
Dear,
Normally, the issue was resolved for G4.11.3. Can you try with this version ?
Thank you, but here comes another problem for me. When I use SBS, the simulation time can last maximum over 1000 picoseconds (normally last hundreds of picosecond), while use IRT , the simulation time normal only last 10-100 picoseconds. So SBS will give much more reaction information than IRT, this is not correct, right? The beam is 1 MeV proton, irradiated to a box with length,width, and height all less than 50 nanometer. Do I need to use G4Scheduler SetEndTime to force it stop at 100 picosecond?
Dear,
No, it is not correct. They should have a very similar result.
Any possible conditions can cause the differences? ![]()
Dear,
I tried with chem 6 example, I set tracking verbose 0, scheduler verbose 2, and change scheduler/endTime from 1 microscond to 10 microscond, then I notice the SBS method show reactions on microsecond level, but IRT method only have reactions below 1 microsecond (20 e- with 100 keV). So I guess the phenomenon is universal, and need to restrict by G4Scheduler SetEndTime. It’s just 100 picsecond may be a harsh limit in my condition, but thanks anyway!!!
dear,
It is true. This because IRT is limited the distance to find nearest molecules up to 1 us.