Geant4 Version: 11.1.1
Operating System: Windows 7 Professional
Compiler/Version: Visual Studio 2022
CMake Version: 3.25.3
I have noticed when using DNA Physics (G4EmDNAPhysics_option2) and Radioactive Decay (G4RadioactiveDecayPhysics), that the relaxation of certain “long”-lived excited daughter isomer are not simulated, but when using Standard Physics (G4EmStandardPhysics_option3) it is.
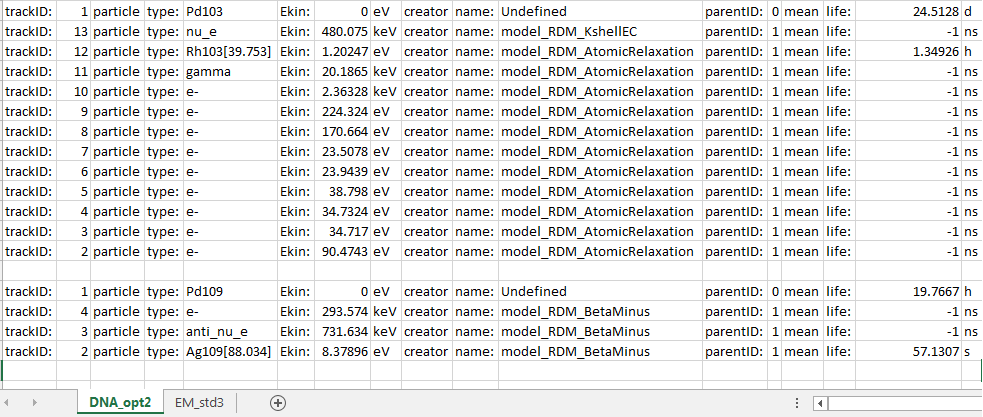
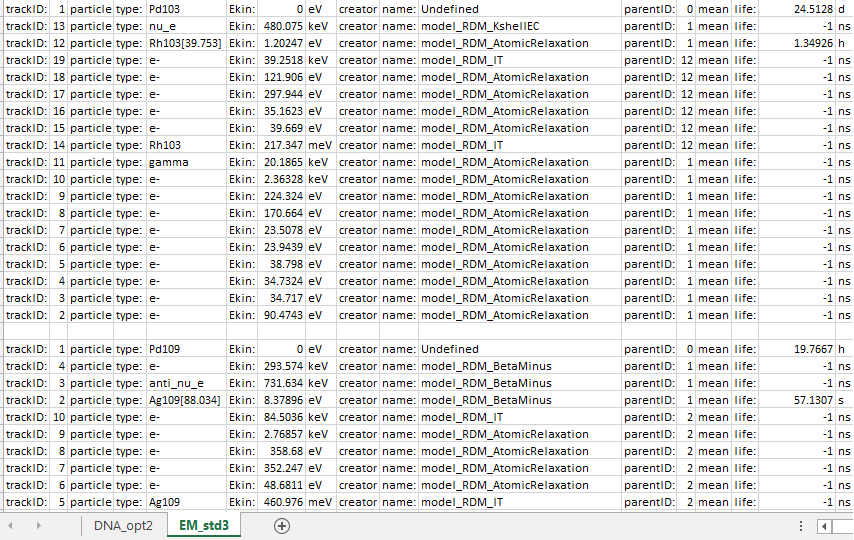
For example the decay of Pd-103 (-> Rh-103* (mean life = 1.35 h) → Rh-103) and Pd-109 (-> Ag-109* (57.13 s) → Ag-109) stop at their excited daughters* but Ra-219 (-> Rn219* (0 ps) → Rn-219 (22.2 ns) → Rn-219 (5.7 s)) continues to Rn-219 using DNA_opt2, but when using EM_Std3 both Pd-103 and Pd-109 continue to their stable products.
I can force the instantaneous decay of “longer” lived particles in DNA by increasing SetThresholdOfHalfLife of the G4NuclideTable (e.g. > 1.35 h) but this will cause them to decay in place, which is not the same scenario as when they were to travel away from their creation site before they relax. (both DNA and EM simulations were done in a G4_Galactic sphere with radius = 5 cm). Is there a life time threshold in DNA Physics which prevents the relaxation of “longer” lived isomers, or a process in EM which is not declared in the modular DNA physics lists?
Regards
Hein