I am trying to read a corsika root file in Geant4, but I have not been successful yet. This is possible?
I think you must use external ROOT libraries
Hi, thanks for your response. What you are trying to tell me, is that I should use the g4root.hh library? Or another one? I imagine that for this I must have rooted root, right?
The g4root.hh header file is used to fill ROOT histograms in a Geant4 simulation, which can then be viewed and analysed outside Geant4, using ROOT. This can be done in a terminal:
> root -l filename.root
See what histograms are available with .ls and view histograms with:
h1->Draw("HIST")
where h1 is the name of a 1D histogram.
Having used Geant4 on and off for a few years, nothing I have seen indicates that viewing ROOT files from within Geant4 is possible, although I would be happy to be corrected if someone knows otherwise.
@emma is correct: the Geant4 toolkit (libraires) does not itself link against ROOT. The g4root utilities in g4analysis provide way to write out a binary-compatible file which can be read by ROOT, without making use of any ROOT library classes.
If you would like to write your application to read in a ROOT file, you will need to write that code yourself, and modify your application-level Makefile (or CMakeLists.txt) to link against the ROOT installation on your system.
Hi @emma and @mkelsey, from what I understand it is very complicated to read a .root file in Geant4, right? But is it possible to read text files?
I ask this, since they made the comment that the .root files can be converted to text files.
Thank you very much for your answers!
Hi @PedroValenciaMex. What exactly is it you are trying to do with this ROOT or text file?
Text files (actually *.mac files) can be read into Geant4 simulations to input initial radiation energies (and other information); I’m not aware of any other situations in which plain text files are used in Geant4. Again, I’m happy to be corrected if I’m wrong.
ROOT files can indeed be converted to text files; I’ve used this to convert a 1D histogram to text in the past.
It’s not complicated to read ROOT files in Geant4, but does require use of the appropriate ROOT types in the application code, together with linking the ROOT libraries for those types to the application.
As noted by @Emma, if you can tell us where in Geant4 you need to read the file from (I’m guessing in one of the User Action classes?), and what sort of ROOT objects it provides, we can provide clearer advice.
Hi, @emma, @bmorgan, an apology for my late reply, I appreciate the inconvenience you have taken to respond to this post.
What I want to do and what I have been breaking my head in recent weeks is: that I can live a simulation of an atmospheric shower made in CORSIKA together with a detector that I am doing in GEANT4.
That is, that the initial particles are obtained from a CORSIKA .root file, and that my GEANT detector reads that file.
I thank you again.
Posting here, as this came up when I was looking for answers on how to use ROOT libraries from my GEANT code.
Caveat emptor: I am not an expert on any of this stuff, and I expect there’s a nice simple way to get this to work. Hopefully someone will post it, or maybe even document it.
In principle it’s easy, that’s what make variables (a.k.a. macros) are for, specifically the LDLIBS variable. And how do you know what to put there? Also easy, since that’s what root-config is for. So ideally, you’d put into your makefile, just before the binmake.gmk line something like
LDLIBS += root-config --libs`
or if you prefer
LDLIBS += $(shell root-config --libs)
and it should work.
If you really want to be organized, you can also put
LDFLAGS = root-config --ldflags`
or so.
But no.
I have no idea why the LDFLAGS line (in my case the only flag is -m64) confused GEANT’s make, but everything works without it, so I let that go.
A bigger problem is that when binmake.gmk sees that the user set up the LDLIBS, it omits some of the libraries that is normally adds to LDLIBS. And those are needed, so the link will fail.
Things almost work if you put the libraries from root-config into LDFLAGS, except that now your library will appear after the ROOT libraries, which means that the linker will not use the ROOT libraries for your library, So you can use ROOT in your main file, but not elsewhere.
Eventually, I just hacked binmake.gmk to accept a variable to be placed after its LDLIBS.
So in lines 418 and 423 of binmake.gmk I now have:
$(LDLIBS) $(AMNONS_2020_HACK)
and in my makefile
AMNONS_2020_HACK = $(shell root-config --libs)
Yet another caveat is that some ROOT headers clash with CLHEP headers. Specifically, TString (which is used everywhere in ROOT) uses “s”, which CLHEP uses for seconds. So I try to (a) avoid ROOT headers in my GEANT headers, (b) in the .cc files, load the ROOT files like this:
#pragma GCC diagnostic push
#pragma GCC diagnostic ignored “-Wshadow”
#include “TMath.h”
#include “TFile.h”
// and any other ROOT headers you need
#pragma GCC diagnostic pop
cheers,
Amnon
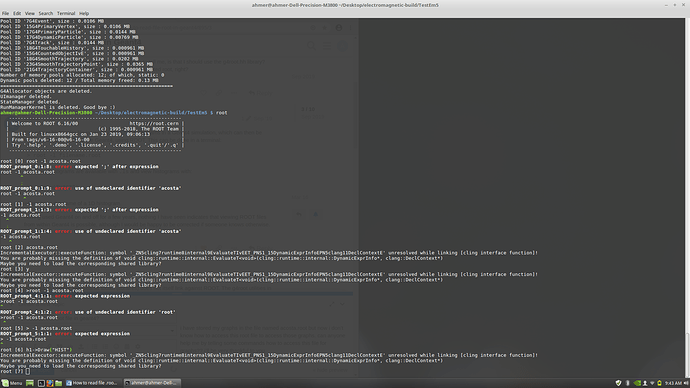
this is the error which came when i used these commands for reading the root file in which my graphs are stored. can you please explain what’s the issue and how can i resolve it?
You should enter into your terminal
root acosta.root
instead of
root
This will execute ROOT and open the file acosta.root.
Or, instead, after executing ROOT with
root
you may open the file typing into ROOT prompt:
TFile file("acosta.root")
Please consult ROOT documentation.
very helpful thanks dear for your time.
can you help me more sorry i’m not that fluent in this work you can say new. 
i wanna ask now how to normalize the two graphs or histograms which i already have with in the acosta.root file?
It’s best to consult the ROOT Documentation, or ask a question on their forum if you can’t find the answer there.
i have the documentation dear but issue is. i didn’t understand how to apply to my histograms to normalize them.
As @bmorgan said, you’re asking questions about how to use ROOT. If you can’t find “histogram normalization” in the documentation, you’re much more likely to get useful answers in the ROOT forums.
Hi, I am trying to use CORSIKA to get the cosmic ray muon flux. Do you have any tutorial of CORSIKA? Thank you very much!