Hello
Could you please help me. I need to obtain energy distribution in a layer of liquid water (layer of “G4_WATER” material with 5-20 micrometers thickness) penetrated by the beam of protons with energy up to 400 MeV at nanoscale (on 10-30 nm voxels) as precise as possible.
When the energy of protons is lower than 100 MeV I can use DNA models which (I assume) are the most accurate models available in Geant 4.10.5. But these models cannot be used for energies higher than 100 MeV and that’s where the problem starts. I tried to use combination of DNA and Livermore models by taking physics list from “microdosimetry” example and replacing standard models with models from “G4EmLivermorePhysics” constructor (which seems to be the most accurate after DNA-models-based constructors). Part of PhysicsList file that describes processes for protons:
//G4StepLimiter* fStepLimiter = new G4StepLimiter();
//pmanager->AddDiscreteProcess(fStepLimiter);
G4hMultipleScattering* msc = new G4hMultipleScattering();
msc->AddEmModel(1,new G4WentzelVIModel());
ph->RegisterProcess(msc, particle);
G4CoulombScattering* pcou = new G4CoulombScattering();
ph->RegisterProcess(pcou, particle);
G4hIonisation* hion = new G4hIonisation();
hion->SetEmModel(new G4BraggModel(), 1);
hion->SetEmModel(new G4BetheBlochModel(), 2);
//hion->SetStepFunction(0.01, 1 * nm);
ph->RegisterProcess(hion, particle);
G4hBremsstrahlung* pb = new G4hBremsstrahlung();
ph->RegisterProcess(pb, particle);
G4hPairProduction* pp = new G4hPairProduction();
ph->RegisterProcess(pp, particle);
G4NuclearStopping* pnuc = new G4NuclearStopping();
ph->RegisterProcess(pnuc, particle);
/…
//G4DNAExcitation, G4DNAIonisation, G4DNAElastic and G4DNAChargeDecrease described here with models set to G4DummyModel to be activated futher as it is done in “microdosimetry” example
…/
I also tried to use G4StepLimiter with the step size of 1 nm and to use SetStepFunction (see commented lines in code). Range cuts for all particles were set to 1 nm. This physics list also uses “G4ElectronCapture” process from “microdosimetry” example that kills all electrons with energies under 7.4 eV.
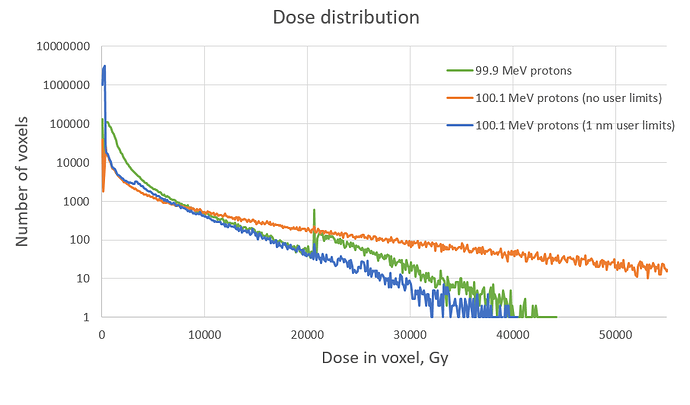
I attached graphs of resulting dose distributions on voxels for protons with the energies of 99.9 MeV (DNA models are used for primary protons) and 100.1 MeV (non-DNA models are used for primary protons). So each point on graphs represents number of voxels that recieved particular dose during simulation. You can see 3 lines corresponding to 3 sets of parameters used:
- initial energy of protons = 99.9 MeV, no user limits used (green line)
- initial energy of protons = 100.1 MeV, no user limits used (red line)
- initial energy of protons = 100.1 MeV, 1 nm user limits used (blue line)
You can see that using user limits (G4StepLimiter) doesn’t solve the problem: green and blue lines should almost match, but they do not. Number of voxels in high doses area differs several times for these two lines. Such result is not acceptable because this area is especially important for our investigation. SetStepFunction on hIoni process did not affect the results at all.
I would appreciate any ideas how to overcome this discrepancy between models for prorons. The